PLAID is a multimodal generative model that simultaneously generates protein 1D sequence and 3D structure, by learning the latent space of protein folding models.
The awarding of the 2024 Nobel Prize to AlphaFold2 marks an important moment of recognition for the of AI role in biology. What comes next after protein folding?
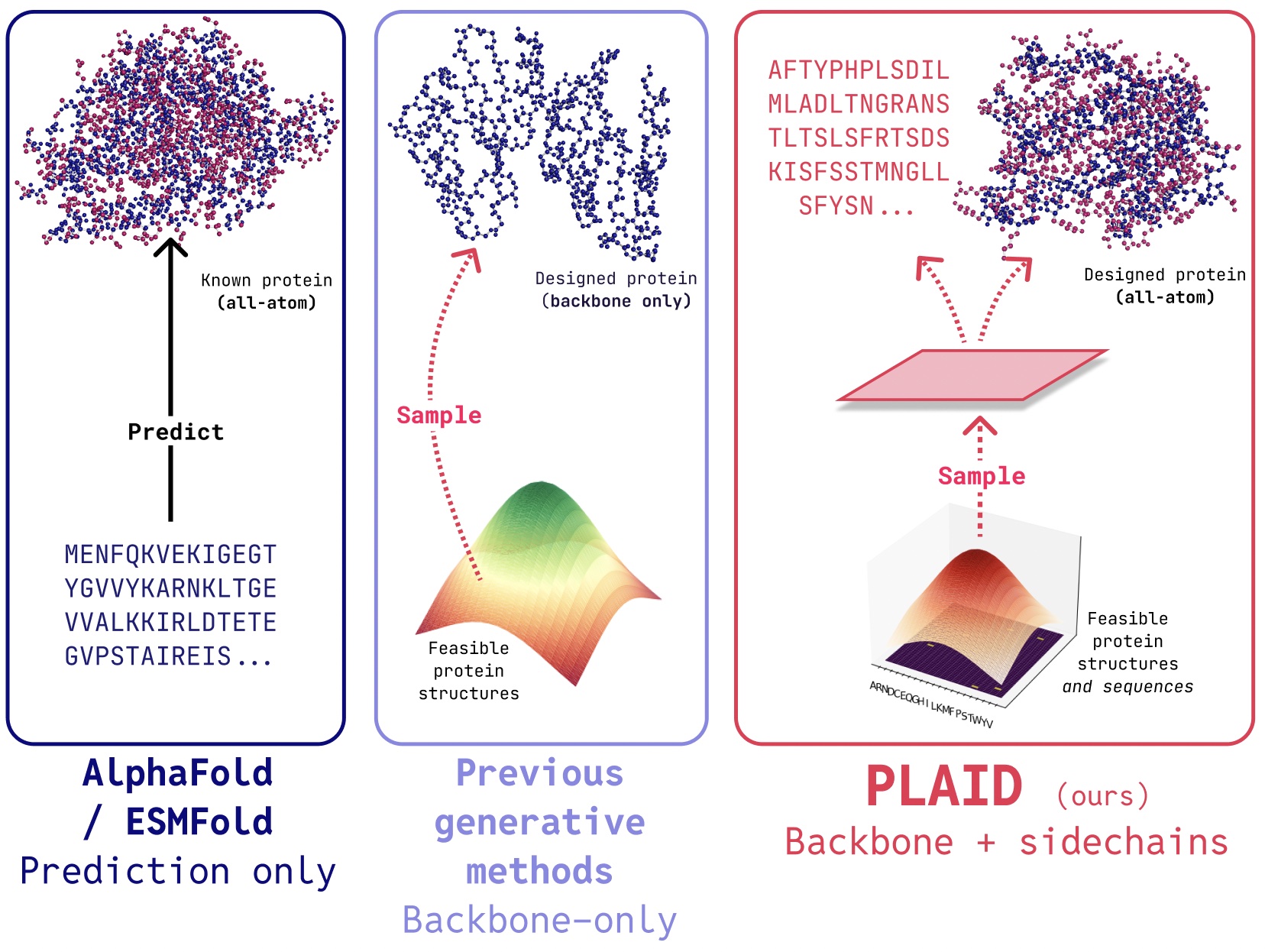
In PLAID, we develop a method that learns to sample from the latent space of protein folding models to generate new proteins. It can accept compositional function and organism prompts, and can be trained on sequence databases, which are 2-4 orders of magnitude larger than structure databases. Unlike many previous protein structure generative models, PLAID addresses the multimodal co-generation problem setting: simultaneously generating both discrete sequence and continuous all-atom structural coordinates.
From structure prediction to real-world drug design
Though recent works demonstrate promise for the ability of diffusion models to generate proteins, there still exist limitations of previous models that make them impractical for real-world applications, such as:
All-atom generation: Many existing generative models only produce the backbone atoms. To produce the all-atom structure and place the sidechain atoms, we need to know the sequence. This creates a multimodal generation problem that requires simultaneous generation of discrete and continuous modalities.
Organism specificity: Proteins biologics intended for human use need to be humanized, to avoid being destroyed by the human immune system.
Control specification: Drug discovery and putting it into the hands of patients is a complex process. How can we specify these complex constraints? For example, even after the biology is tackled, you might decide that tablets are easier to transport than vials, adding a new constraint on soluability.
Generating “useful” proteins
Simply generating proteins is not as useful as controlling the generation to get useful proteins. What might an interface for this look like?

For inspiration, let’s consider how we’d control image generation via compositional textual prompts (example from Liu et al., 2022).
In PLAID, we mirror this interface for control specification. The ultimate goal is to control generation entirely via a textual interface, but here we consider compositional constraints for two axes as a proof-of-concept: function and organism:
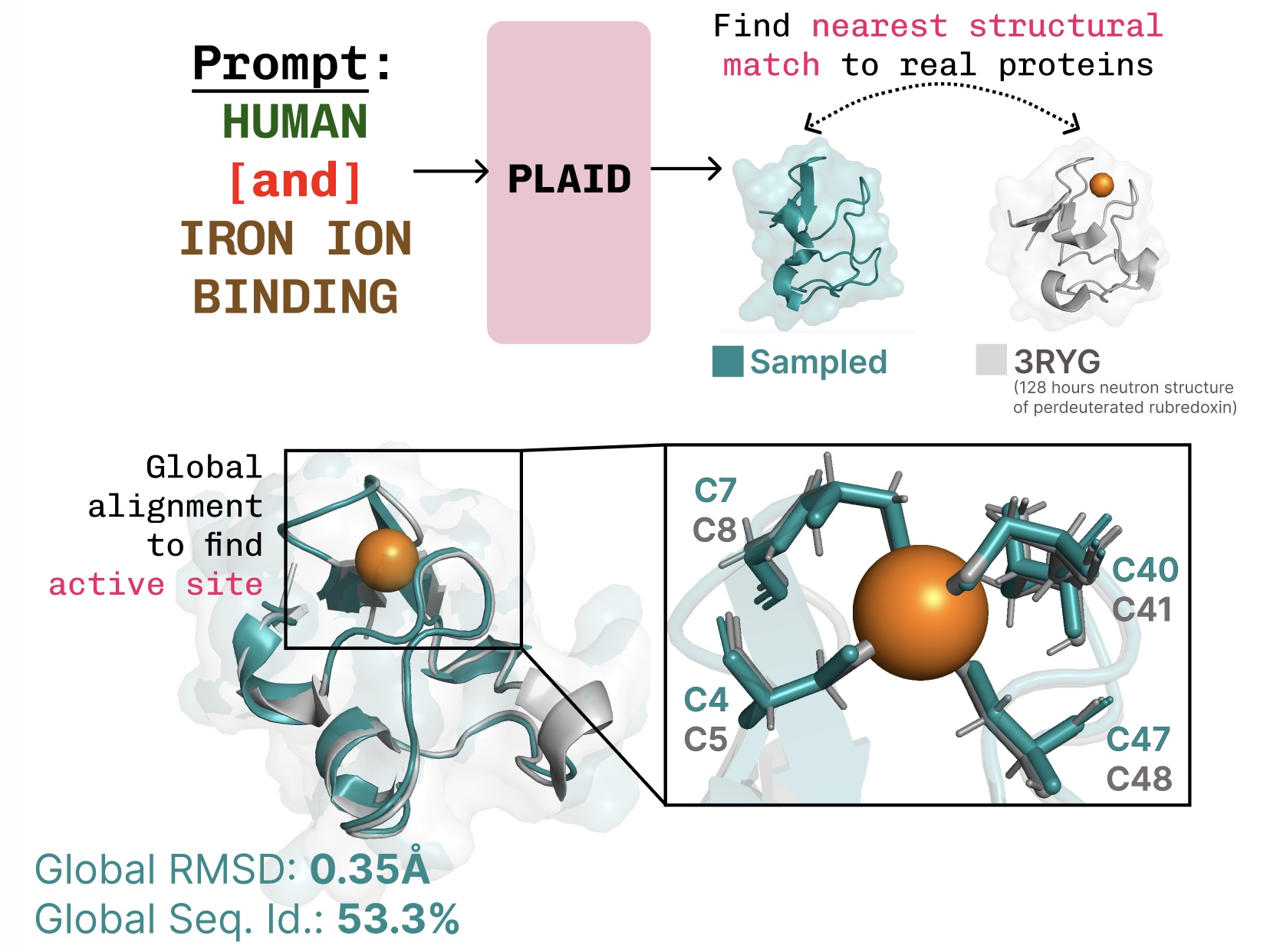
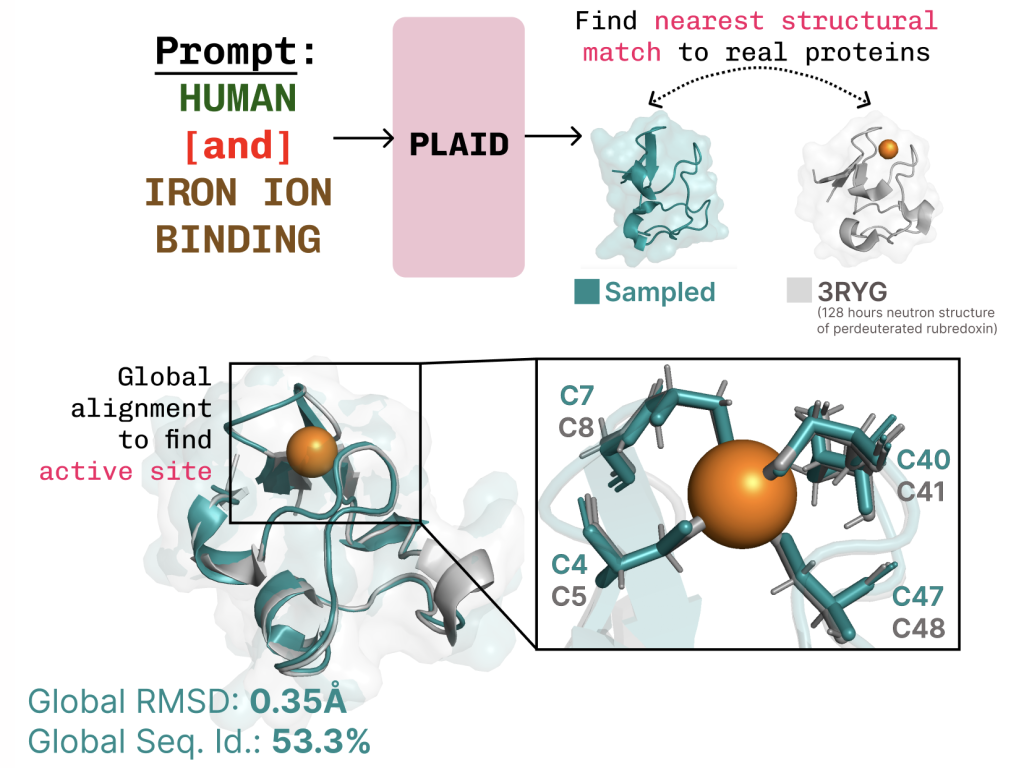
Learning the function-structure-sequence connection. PLAID learns the tetrahedral cysteine-Fe2+/Fe3+ coordination pattern often found in metalloproteins, while maintaining high sequence-level diversity.
Training using sequence-only training data
Another important aspect of the PLAID model is that we only require sequences to train the generative model! Generative models learn the data distribution defined by its training data, and sequence databases are considerably larger than structural ones, since sequences are much cheaper to obtain than experimental structure.
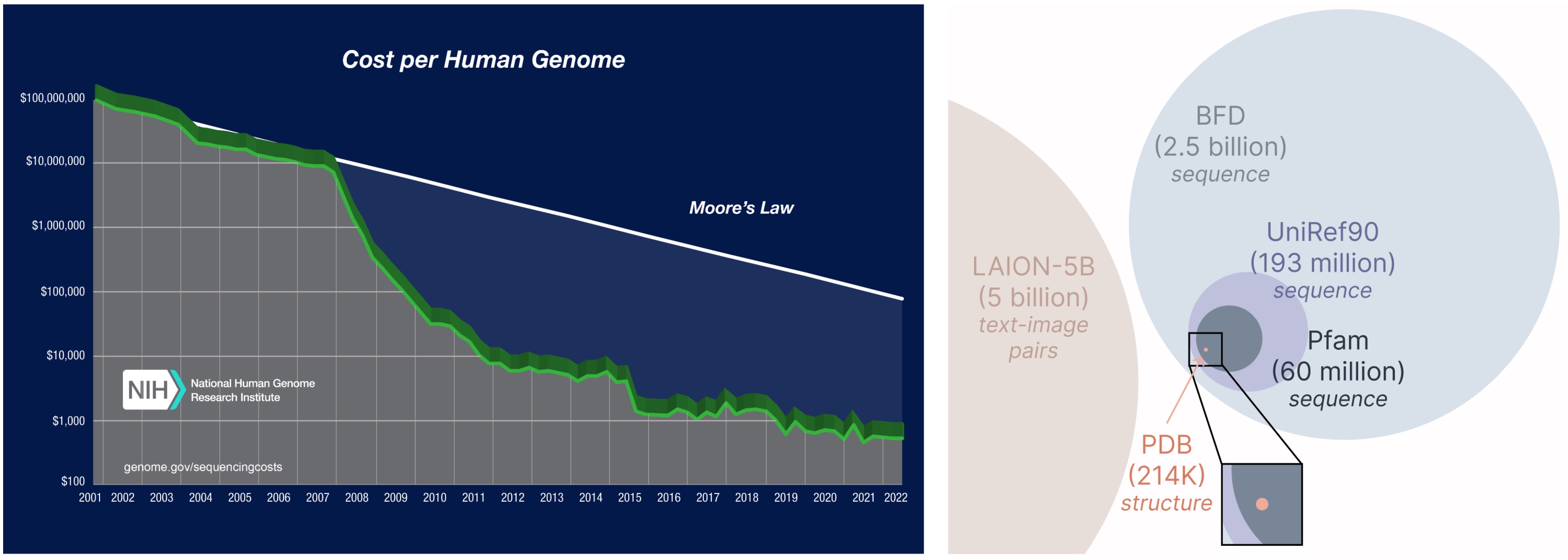
Learning from a larger and broader database. The cost of obtaining protein sequences is much lower than experimentally characterizing structure, and sequence databases are 2-4 orders of magnitude larger than structural ones.
How does it work?
The reason that we’re able to train the generative model to generate structure by only using sequence data is by learning a diffusion model over the latent space of a protein folding model. Then, during inference, after sampling from this latent space of valid proteins, we can take frozen weights from the protein folding model to decode structure. Here, we use ESMFold, a successor to the AlphaFold2 model which replaces a retrieval step with a protein language model.
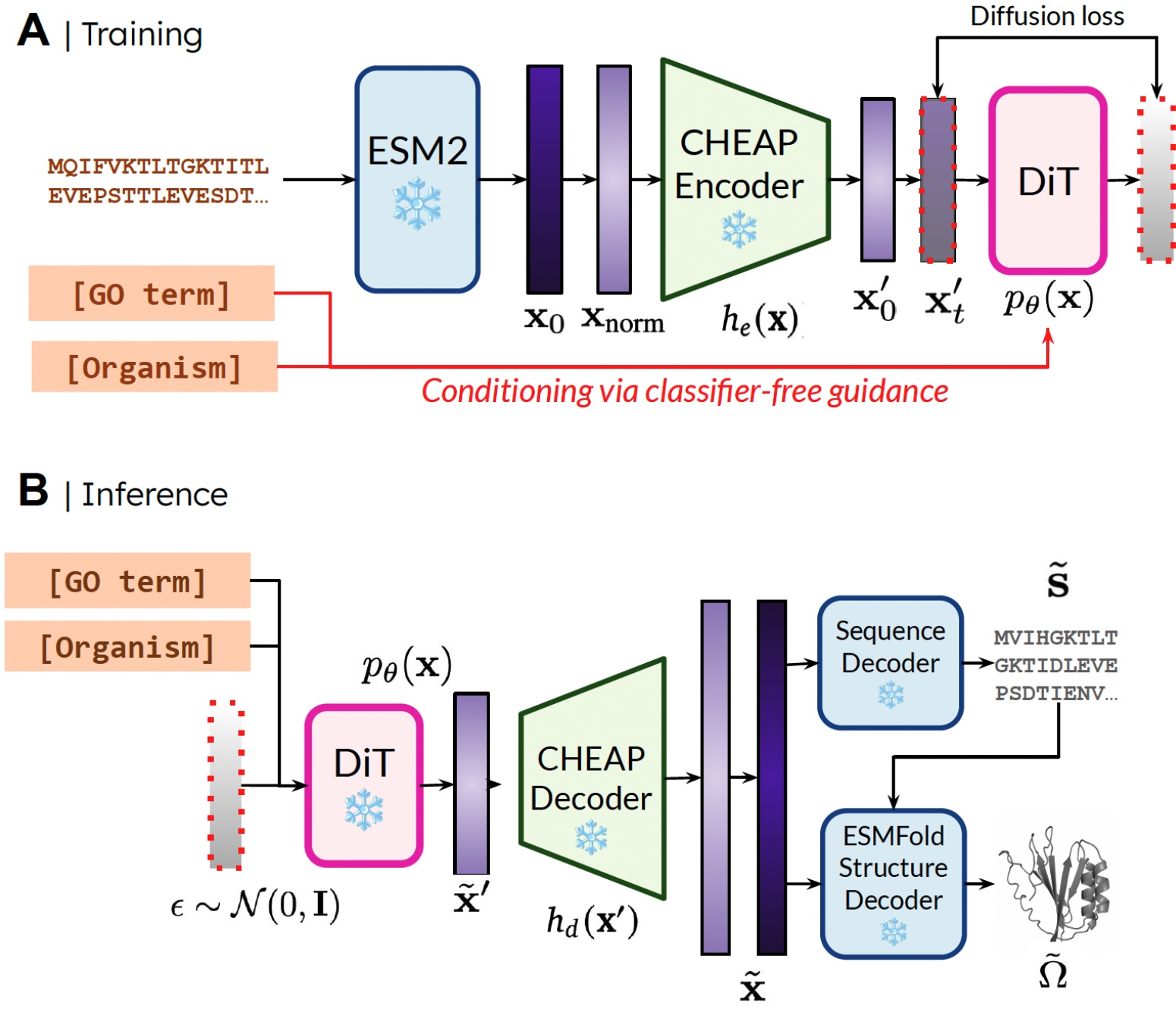
Our method. During training, only sequences are needed to obtain the embedding; during inference, we can decode sequence and structure from the sampled embedding. ❄️ denotes frozen weights.
In this way, we can use structural understanding information in the weights of pretrained protein folding models for the protein design task. This is analogous to how vision-language-action (VLA) models in robotics make use of priors contained in vision-language models (VLMs) trained on internet-scale data to supply perception and reasoning and understanding information.
Compressing the latent space of protein folding models
A small wrinkle with directly applying this method is that the latent space of ESMFold – indeed, the latent space of many transformer-based models – requires a lot of regularization. This space is also very large, so learning this embedding ends up mapping to high-resolution image synthesis.
To address this, we also propose CHEAP (Compressed Hourglass Embedding Adaptations of Proteins), where we learn a compression model for the joint embedding of protein sequence and structure.
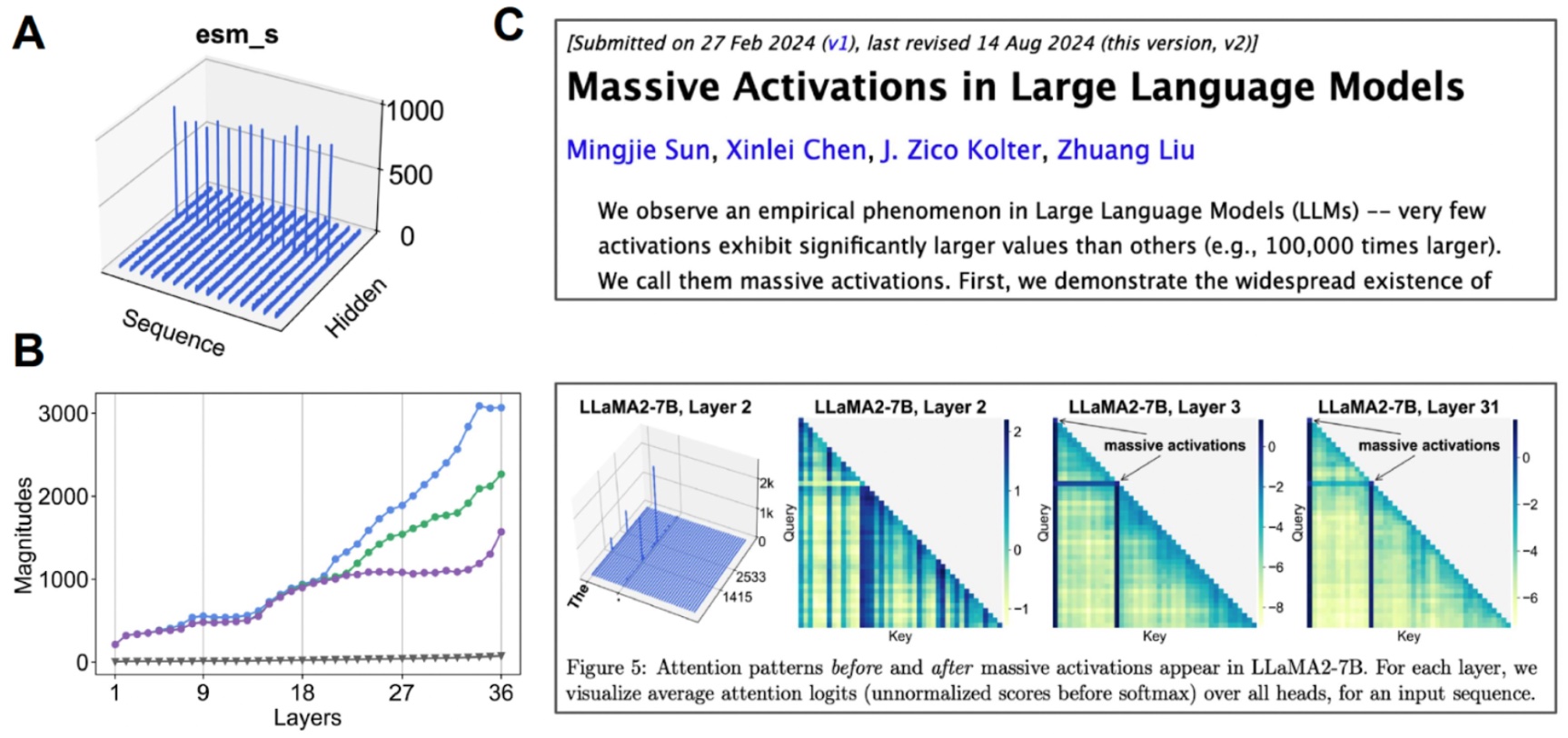
Investigating the latent space. (A) When we visualize the mean value for each channel, some channels exhibit “massive activations”. (B) If we start examining the top-3 activations compared to the median value (gray), we find that this happens over many layers. (C) Massive activations have also been observed for other transformer-based models.
We find that this latent space is actually highly compressible. By doing a bit of mechanistic interpretability to better understand the base model that we are working with, we were able to create an all-atom protein generative model.
What’s next?
Though we examine the case of protein sequence and structure generation in this work, we can adapt this method to perform multi-modal generation for any modalities where there is a predictor from a more abundant modality to a less abundant one. As sequence-to-structure predictors for proteins are beginning to tackle increasingly complex systems (e.g. AlphaFold3 is also able to predict proteins in complex with nucleic acids and molecular ligands), it’s easy to imagine performing multimodal generation over more complex systems using the same method.
If you are interested in collaborating to extend our method, or to test our method in the wet-lab, please reach out!
Further links
If you’ve found our papers useful in your research, please consider using the following BibTeX for PLAID and CHEAP:
title={Generating All-Atom Protein Structure from Sequence-Only Training Data},
author={Lu, Amy X and Yan, Wilson and Robinson, Sarah A and Yang, Kevin K and Gligorijevic, Vladimir and Cho, Kyunghyun and Bonneau, Richard and Abbeel, Pieter and Frey, Nathan},
journal={bioRxiv},
pages={2024–12},
year={2024},
publisher={Cold Spring Harbor Laboratory}
}
title={Tokenized and Continuous Embedding Compressions of Protein Sequence and Structure},
author={Lu, Amy X and Yan, Wilson and Yang, Kevin K and Gligorijevic, Vladimir and Cho, Kyunghyun and Abbeel, Pieter and Bonneau, Richard and Frey, Nathan},
journal={bioRxiv},
pages={2024–08},
year={2024},
publisher={Cold Spring Harbor Laboratory}
}
You can also checkout our preprints (PLAID, CHEAP) and codebases (PLAID, CHEAP).
Some bonus protein generation fun!
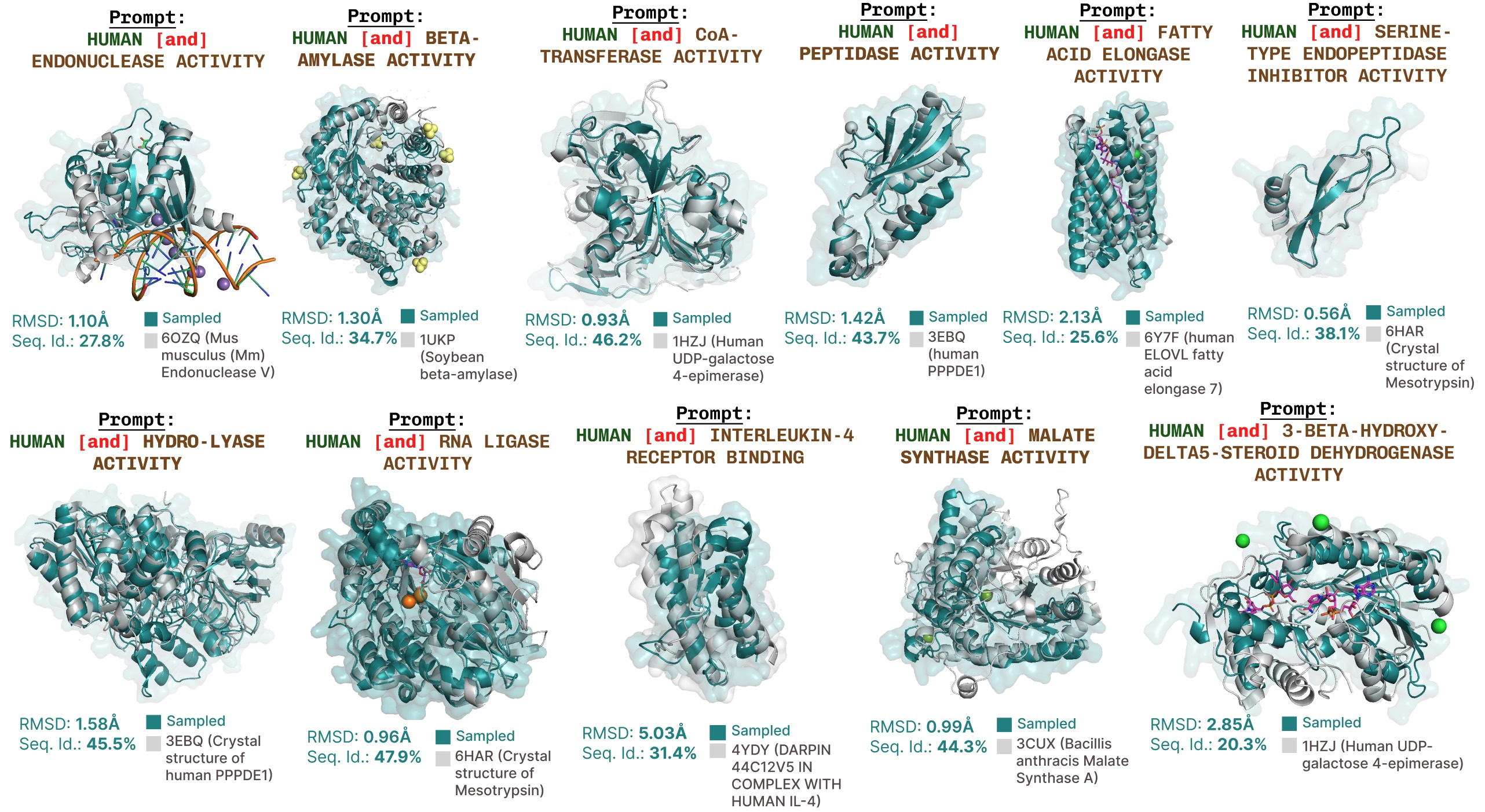
Additional function-prompted generations with PLAID.
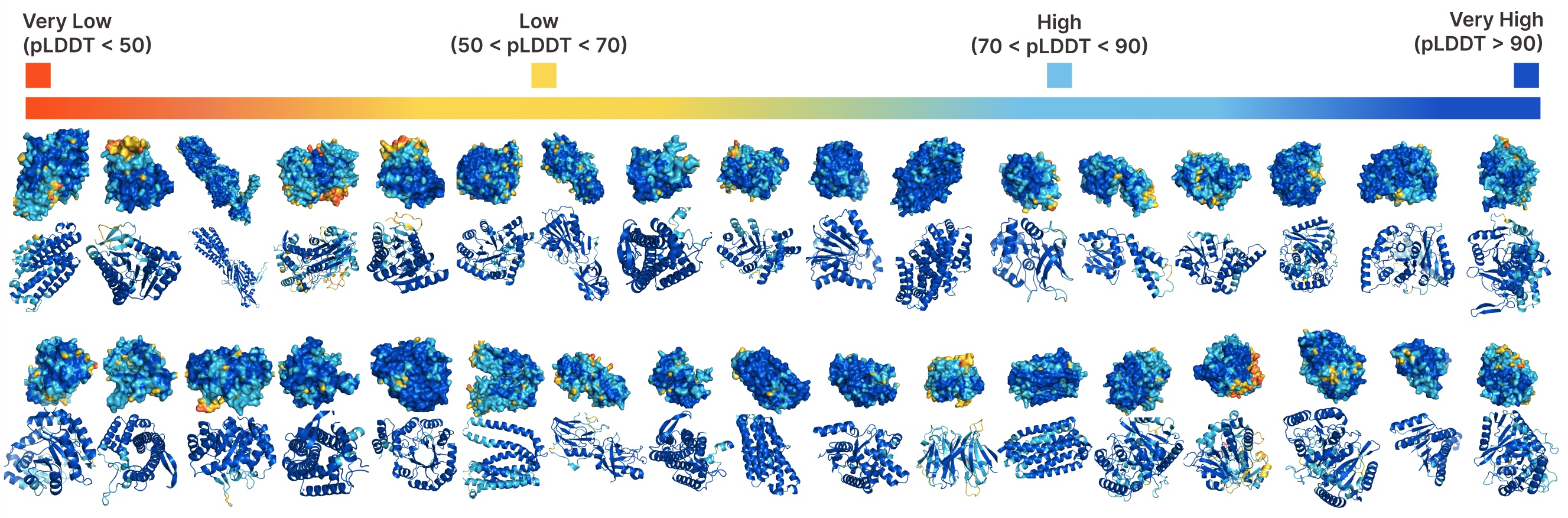
Unconditional generation with PLAID.
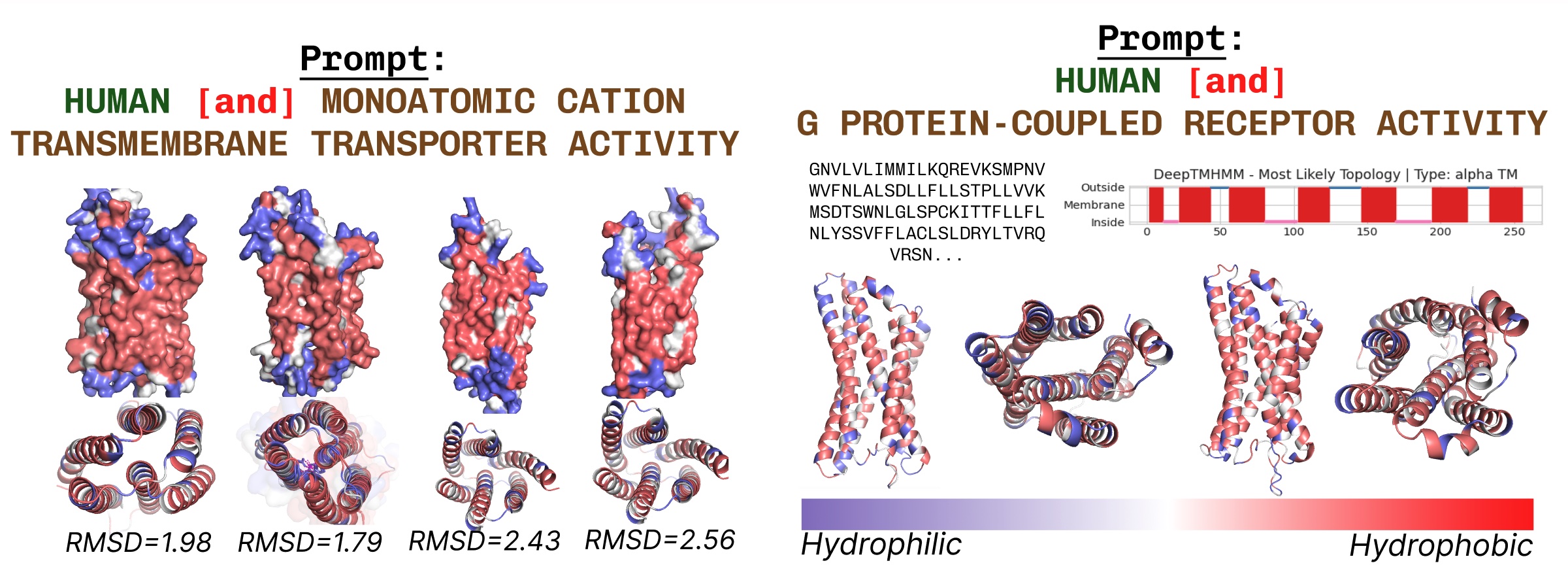
Transmembrane proteins have hydrophobic residues at the core, where it is embedded within the fatty acid layer. These are consistently observed when prompting PLAID with transmembrane protein keywords.
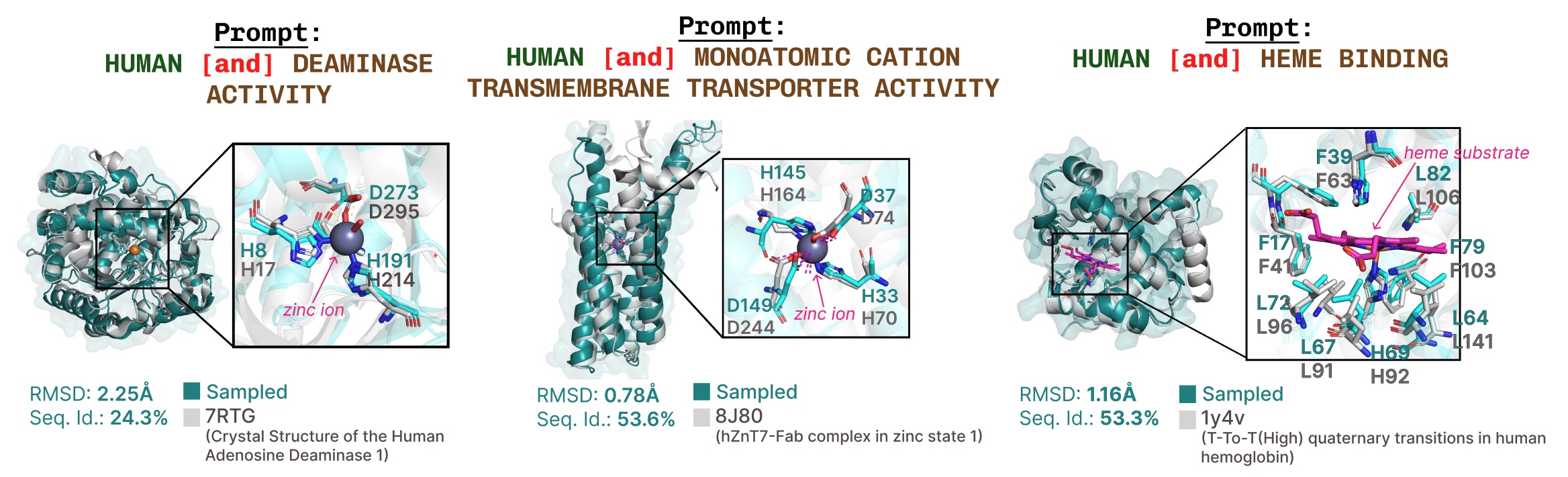
Additional examples of active site recapitulation based on function keyword prompting.
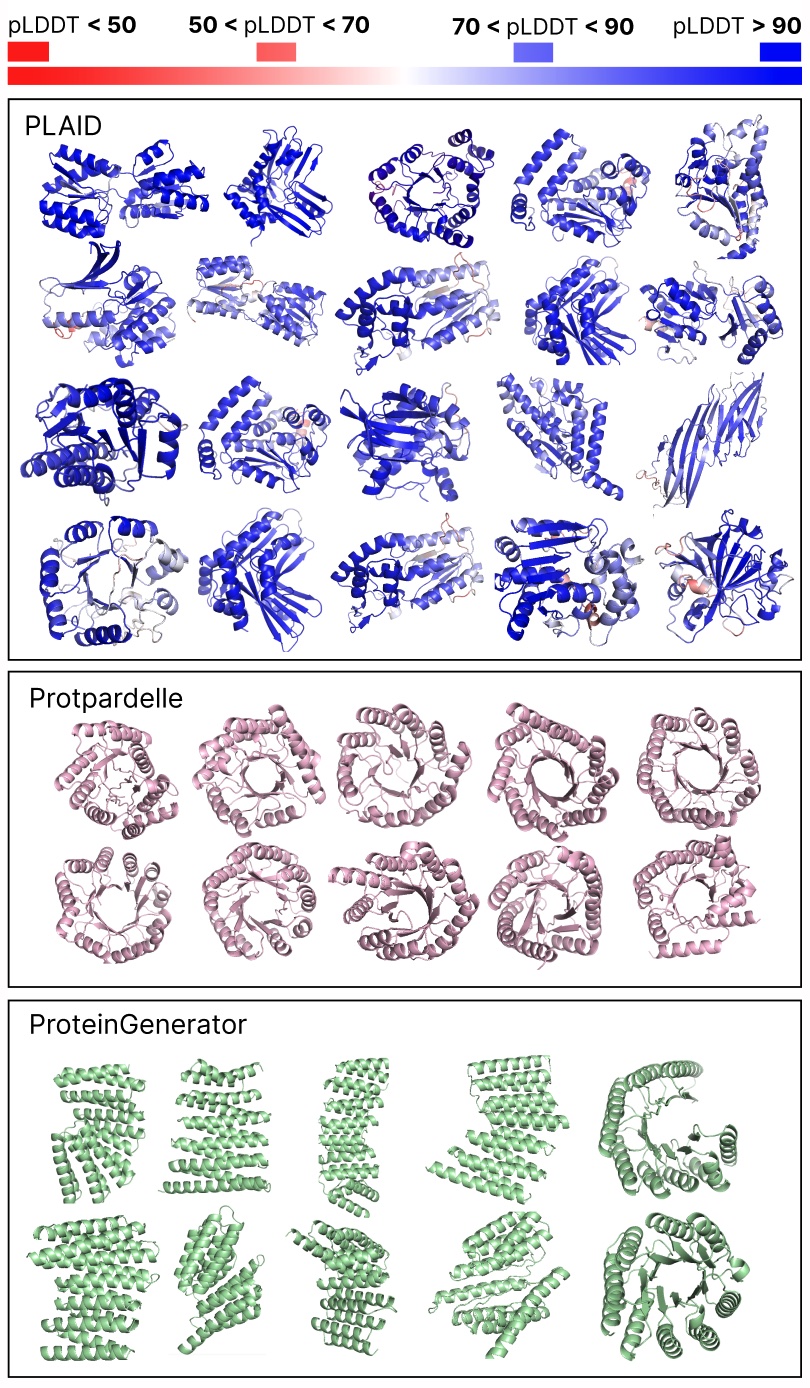
Comparing samples between PLAID and all-atom baselines. PLAID samples have better diversity and captures the beta-strand pattern that has been more difficult for protein generative models to learn.
Acknowledgements
Thanks to Nathan Frey for detailed feedback on this article, and to co-authors across BAIR, Genentech, Microsoft Research, and New York University: Wilson Yan, Sarah A. Robinson, Simon Kelow, Kevin K. Yang, Vladimir Gligorijevic, Kyunghyun Cho, Richard Bonneau, Pieter Abbeel, and Nathan C. Frey.